2.2. Canonical Ensemble#
Overview#
In a closed system—one that exchanges energy but not matter with its surroundings—the appropriate statistical description is the canonical ensemble.
Show code cell source
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
from myst_nb import glue
# Helper function to plot a system
def plot_system(ax, title, annotations, boundary_color='b'):
box = mpatches.FancyBboxPatch((0, 0), 1, 1, boxstyle='roundtooth', ec=boundary_color, fc='w')
ax.add_patch(box)
ax.set_title(title, fontsize=14)
ax.text(0.5, 0.5, 'System', ha='center', va='center', fontsize=12)
ax.text(0.5, -0.65, 'Surroundings', ha='center', va='center', fontsize=12)
ax.text(0.5, 1.3, 'Boundary', ha='center', va='bottom', fontsize=12, color=boundary_color)
for annotation in annotations:
if "arrowprops" in annotation: # Arrow annotations
ax.annotate('', **annotation)
else: # Text annotations
ax.text(**annotation)
ax.set_xlim(-1, 2)
ax.set_ylim(-1, 2)
ax.set_aspect('equal')
ax.axis('off')
# Define annotations for each system
annotations = [
dict(xy=(-0.6, 0.15), xytext=(0.15, 0.15), arrowprops=dict(arrowstyle='<->', color='r')),
dict(x=-1, y=0.3, s='Energy', ha='left', va='bottom', fontsize=12, color='r'),
]
fig, ax = plt.subplots(1, 1, figsize=(4, 4))
plot_system(ax, "", annotations)
glue('closed-system', fig, display=False)
plt.close(fig)
Fig. 13 A closed system, exchanging energy but not matter with its surroundings.#
Probability of a Microstate in the Canonical Ensemble#
Consider an ensemble of \(\mathcal{A}\) closed systems exchanging energy with a heat bath at temperature \(T\). A heat bath is an environment that can absorb or release energy without changing its temperature because it is much larger than the system.
How Can an Environment Absorb or Release Energy Without Changing Its Temperature?
The key point is that the temperature of a collection of particles is determined by the average kinetic energy per particle. According to the equipartition theorem, for particles with three translational degrees of freedom the total kinetic energy is
Thus, for a fixed amount of energy transfer, the corresponding change in temperature is given by
Because the environment has many more particles than the system (\(N_\text{env} \gg N_\text{sys}\)), the same amount of energy \(\Delta E\) will result in a much smaller \(\Delta T\) for the environment. In our example, even though the system (with \(N_\text{sys} = 6.022\times10^{23}\) particles) absorbs energy and its temperature increases from 300 K to 400 K, the environment (with \(N_\text{env} = 6.022\times10^{24}\) particles, i.e. 10 times as many) loses the same amount of energy per mole but its temperature only drops from 400 K to 390 K.
This illustrates that an environment (or heat bath) can absorb or release energy almost isothermally because its very large number of particles (and hence its high heat capacity) means that the fractional change in average kinetic energy per particle is negligible—even though energy is being exchanged.
Thus, even though the total kinetic energy of the universe (system plus environment) remains constant, the environment’s temperature barely changes because any energy loss or gain is diluted among a huge number of particles.
Ratio of Numbers of Systems in Two Microstates is a Function of Their Relative Energies#
Intuition tells us that a system is more likely to be found in microstates with lower energy.
Building This Intuition
Consider a raindrop that can exist at two different elevations: one high up at Lake Itasca in northern Minnesota and one low down in the Mississippi River in St. Louis. At the higher elevation, the raindrop has more gravitational potential energy, making that state less favorable. Consequently, the raindrop is more likely to be found at the lower elevation, where its gravitational potential energy is reduced. In essence, systems tend to “prefer” lower energy states, which is why, on a macroscopic scale, water naturally flows downhill.
Mathematically, this intuition asserts that the ratio of the numbers \(a_1\) and \(a_2\) of systems in two microstates 1 and 2 is given by
where \(E_1\) and \(E_2\) are the energies of the two microstates. The function \(f\) depends only on the difference in energy between the two states.
Why Does the Ratio Depend Only on the Energy Difference?
The energy of a system is defined relative to an arbitrary (but often convenient) reference level. For example,
The kinetic energy of a moving particle is defined relative to a stationary particle.
The gravitational potential energy of a raindrop is defined relative to the surface of the Earth.
Therefore, the absolute energy of a system is not essential in determining the likelihood of finding the system in a particular microstate, only the difference in energy between two microstates.
Finding an Acceptable Form for \(f\)#
Since \(\{ a_1, a_2, a_3, \ldots \}\) is a set of numbers, we can write
If \(f\) is “well-behaved” (i.e., continuous, measurable, etc.), it must be of the form
where \(\beta\) is an undetermined constant, which we will determine later to be \(\frac{1}{k_\text{B} T}\).
Checking the Form of \(f\)
Using this form for \(f\), we can verify that
Converting \(f\) to a Probability#
Separating the variables with indices \(m\) and \(n\),
where \(\mathcal{C}\) is a constant. Therefore, the number of systems in a microstate with energy \(E_m\) is
The constant \(\mathcal{C}\) is determined by the normalization condition
Solving the second equation for \(\mathcal{C}\) and substituting it into the equation for \(a_m\) gives
where \(p_m\) is the probability of finding the system in microstate \(m\) and \(Q\) is called the partition function, which is the essential quantity in statistical mechanics.
Two-State System#
Consider a system with two states: state 1 with energy \(E_1\) and state 2 with energy \(E_2\).
Chemical Context |
State 1 |
State 2 |
|---|---|---|
Electronic transitions in atoms or molecules |
Ground state |
Excited state |
Donor–acceptor electron transfer |
Reduced state |
Oxidized state |
Molecular isomerization |
Reactant |
Product |
Defects in solids |
Defect-free |
Defective |
Protein folding |
Unfolded |
Folded |
Partition Function for a Two-State System#
The partition function \(Q_\text{two-state}\) for a two-state system is
where \(\Delta E = E_2 - E_1\) is the energy difference between the two states.
Probability of Finding the System in State 1#
The probability of finding the system in state 1 is
Probability of Finding the System in State 2#
The probability of finding the system in state 2 is
Partition Function as the Effective Number of Thermally Accessible Microstates#
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
from scipy.constants import k, eV
from labellines import labelLines
from myst_nb import glue
from matplotlib.patches import Rectangle
k_B = k / eV # Boltzmann constant in eV/K
# Define the partition function for a two-state system
def partition_function_two_state(E1, E2, T):
beta = 1 / (k_B * T)
return np.exp(-beta * E1) + np.exp(-beta * E2)
# Calculate the partition function for a two-state system
E1 = 0
E2 = 0.01 # Energy difference between the two states in eV
T_values = np.linspace(1, 1000, 1000)
Q_values = [partition_function_two_state(E1, E2, T) for T in T_values]
# Calculate the probabilities of finding the system in each state for a two-state system
p1_values = [np.exp(-1 / (k_B * T) * E1) / Q for T, Q in zip(T_values, Q_values)]
p2_values = [np.exp(-1 / (k_B * T) * E2) / Q for T, Q in zip(T_values, Q_values)]
# Plot the partition function and the probabilities of finding the system in each state
fig, axs = plt.subplots(1, 2, figsize=(8, 4))
# Plot the partition function on axs[0]
axs[0].plot(T_values, Q_values, 'k-')
axs[0].set_xlabel('Temperature (K)')
axs[0].set_ylabel('$Q_{\\text{two-state}}$')
axs[0].grid(True)
axs[0].annotate(
'$\\rightarrow 1$ accessible\nmicrostate', xy=(40, Q_values[0] + 0.01), xytext=(300, 1.1),
arrowprops=dict(arrowstyle='->', color='b'),
bbox=dict(boxstyle='round,pad=0.3', fc='w', ec='b'),
ha='center', va='center', color='b'
)
axs[0].annotate(
'$\\rightarrow 2$ accessible\nmicrostates', xy=(T_values[-1], Q_values[-1]), xytext=(750, 1.7),
arrowprops=dict(arrowstyle='->', color='m'),
bbox=dict(boxstyle='round,pad=0.3', fc='w', ec='m'),
ha='center', va='center', color='m'
)
axs[0].set_xlim(0, 1000)
axs[0].set_ylim(1, 2)
# Plot the probabilities of finding the system in each state on axs[1]
p1_line, = axs[1].plot(T_values, p1_values, 'b-', label='State 1')
p2_line, = axs[1].plot(T_values, p2_values, 'r-', label='State 2')
labelLines([p1_line, p2_line], zorder=2.5)
axs[1].set_xlabel('Temperature (K)')
axs[1].set_ylabel('Probability')
axs[1].grid(True)
axs[1].set_ylim(0, 1) # ensure y-axis spans from 0 to 1
# Add tall outlined rectangles around the probabilities at low and high temperatures.
# For T -> 0: highlight T from 1 to 50 K.
# For T = 1000: highlight T from 950 to 1000 K.
rect_low = Rectangle((1, 0), 50 - 1, 1, edgecolor='b', facecolor='b', linestyle='-', alpha=0.2)
rect_high = Rectangle((950, 0), 1000 - 950, 1, edgecolor='m', facecolor='m', linestyle='-', alpha=0.2)
axs[1].add_patch(rect_low)
axs[1].add_patch(rect_high)
axs[1].annotate(
'Only state 1 is accessed', xy=(50, 0.95), xytext=(100, 0.95),
arrowprops=dict(arrowstyle='-', color='b'),
ha='left', va='center', color='b'
)
axs[1].annotate(
'Both states are accessed', xy=(950, 0.05), xytext=(900, 0.05),
arrowprops=dict(arrowstyle='-', color='m'),
ha='right', va='center', color='m'
)
plt.tight_layout()
glue('two-state-system', fig, display=False)
plt.close(fig)
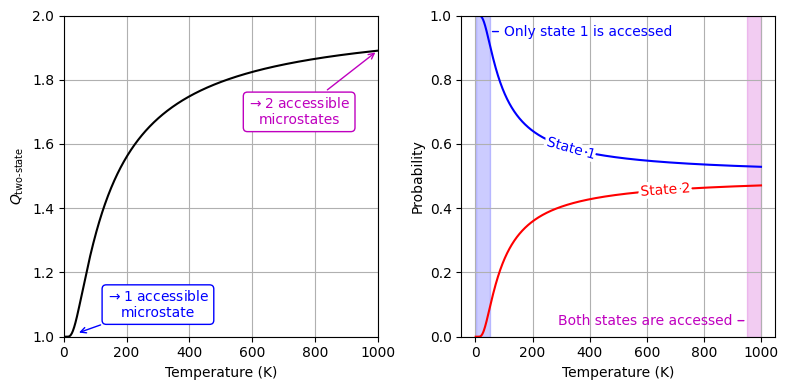
Fig. 14 Partition function of a two-state system and the probabilities of finding it in each state as a function of temperature. The energy difference between the two states is 0.01 eV.#
