2.6. Harmonic Oscillator#
Overview#
This section discusses the quantum mechanical treatment of a harmonic oscillator, along with the derivation of key thermodynamic properties using the partition function. We begin with a review of the one-dimensional case, generalize to three dimensions, and then show how the classical partition function emerges in the high-temperature limit.
Review of the One-Dimensional Harmonic Oscillator#
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d.art3d import Poly3DCollection
from scipy.constants import k, eV
from scipy.special import hermite, factorial
from labellines import labelLines
from myst_nb import glue
from numpy.linalg import eigh
fig, axs = plt.subplot_mosaic([[0]], figsize=(4, 4))
def harmonic_potential(x, m, omega):
"""
Calculates the potential matrix for a particle in a one-dimensional harmonic potential
Parameters
----------
x : array-like
The positions of the particles.
m : float
The mass of the particle.
omega : float
The frequency of the harmonic potential
Returns
-------
array
The potential matrix.
"""
V = 0.5 * m * omega**2 * x**2
V_matrix = np.diag(V)
return V_matrix
def off_diagonal_identity_matrix(n_points):
matrix = np.zeros((n_points, n_points))
for i in range(n_points):
if i == 0:
matrix[i + 1, i] = 1
elif i == n_points - 1:
matrix[i - 1, i] = 1
else:
matrix[i - 1, i] = 1
matrix[i + 1, i] = 1
return matrix
def laplacian_matrix(x):
"""
Calculates the Laplacian matrix for a particle in a one-dimensional potential.
Parameters
----------
x : array-like
The positions of the particles.
Returns
-------
array
The Laplacian matrix.
"""
Delta_x = x[1] - x[0]
n_points = len(x)
off_diag = off_diagonal_identity_matrix(n_points)
Laplacian = (1 / (Delta_x**2)) * (-2 * np.eye(n_points) + off_diag)
return Laplacian
# Define constants in atomic units
hbar = 1
m = 1
# Harmonic potential
omega = 1
# Discretize the system
n_points = 2000
L = 40
x = np.linspace(-L/2, L/2, n_points)
# Construct the Hamiltonian matrix
H_harm = -hbar**2 / (2 * m) * laplacian_matrix(x) + harmonic_potential(x, m=1, omega=1)
# Solve for eigenvalues and eigenfunctions
E_harm, psi_harm = np.linalg.eigh(H_harm)
# Plot a one-dimensional quadratic potential
axs[0].plot(x, np.diag(harmonic_potential(x, m, omega)), "k-", zorder=2.5, lw=4)
# Plot the energy levels (blue lines)
for n in range(4):
energy = axs[0].plot(x, np.ones_like(x) * E_harm[n], color='blue', label=r'$E_{%d}$' % n)
labelLines(energy, xvals=[-L/24], zorder=2.5)
# Plot the wavefunctions (red curves)
for n in range(4):
wavefunction = axs[0].plot(x, psi_harm[:, n] * 4 + E_harm[n], color='red', label=r'$\psi_{%d}$' % n)
labelLines(wavefunction, xvals=[L/24], zorder=2.5)
# Format plot
axs[0].set_xlim(-L/8, L/8)
ymin = np.min(np.diag(harmonic_potential(x, m, omega)))
ymax = np.max(psi_harm[:, -1]) + E_harm[3]
yrange = ymax - ymin
ybuffer = 0.05 * yrange
axs[0].set_ylim(ymin - ybuffer, ymax + ybuffer * 2)
axs[0].set_xlabel("Position")
axs[0].set_ylabel("Energy")
axs[0].set_xticks([])
axs[0].set_yticks([])
axs[0].spines['top'].set_visible(False)
axs[0].spines['right'].set_visible(False)
axs[0].spines['left'].set_visible(False)
axs[0].spines['bottom'].set_visible(False)
plt.tight_layout()
glue("harmonic_oscillator", fig, display=False)
plt.close(fig)
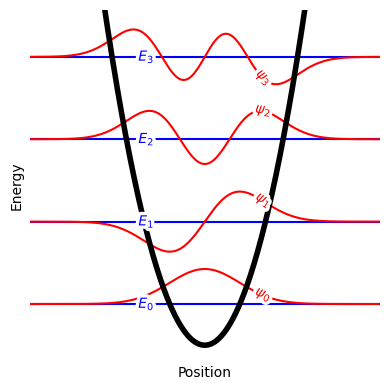
Fig. 17 Enery levels and wavefunctions for a one-dimensional harmonic oscillator. The energy levels \(E_n\) are shown as blue lines, while the wavefunctions \(\psi_n\) are shown as red curves.#
For a one-dimensional harmonic oscillator of frequency \(\omega\), the quantized energy levels are given by
where \(\hbar\) is the reduced Planck constant and \(n\) is a positive integer.
Partition Function for a Harmonic Oscillator#
The partition function for a one-dimensional harmonic oscillator is:
Evaluation of the Sum#
The \(\sum_{n=0}^{\infty} e^{-\beta \hbar \omega n}\) term is a geometric series, which can be evaluated as
Substituting \(x = e^{-\beta \hbar \omega}\) gives:
Is \(|x| < 1\)?
Ensemble Averages for a Harmonic Oscillator#
Natural Logarithm of the Partition Function#
Taking the logarithm of \(q\):
Internal Energy#
The internal energy \(U\) is found via:
Derivative of \(\ln(1 - e^{-\beta \hbar \omega})\)
We often define the vibrational temperature \(\Theta_{\text{vib}}\) as
Hence,
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d.art3d import Poly3DCollection
from scipy.constants import k, eV
from labellines import labelLines
from myst_nb import glue
kB = k / eV # Boltzmann constant in eV/K
fig, axs = plt.subplot_mosaic([[0]], figsize=(4, 4))
Theta_vib = 805 # K, for a Cl2 molecule
hbar_omega = kB * Theta_vib
T = np.linspace(10, 1000, 100)
Tr = T / Theta_vib
U = hbar_omega / 2 + hbar_omega / (np.exp(Theta_vib / T) - 1)
axs[0].plot(Tr, U / hbar_omega, "k-")
axs[0].set_xlabel(r"$T / \Theta_{\text{vib}}$")
axs[0].set_ylabel(r"$U / \left( \hbar \omega \right)$")
zero_point_energy = axs[0].plot([np.min(Tr), np.max(Tr)], [0.5, 0.5], "b--", label="Zero-point energy")
labelLines(zero_point_energy, zorder=2.5)
x = Tr[Tr >= 0.5]
kT = axs[0].plot(x, x, "r--", label=r"$U \rightarrow k_{\text{B}} T$")
labelLines(kT, xvals=[Tr[-1] * 3 / 4], zorder=2.5)
plt.tight_layout()
glue("harmonic_oscillator_energy", fig, display=False)
plt.close(fig)
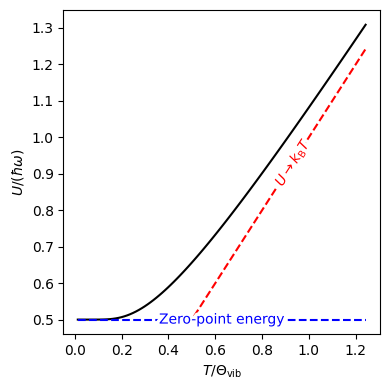
Fig. 18 Internal energy of a harmonic oscillator as a function of temperature. The zero-point energy is shown as a blue dashed line, while the high-temperature limit is shown as a red dashed line.#
Heat Capacity at Constant Volume#
In Section 2.3, we showed that
From Equation (21), it follows that:
Complete Derivation of \(C_V\)
Show code cell source
fig, axs = plt.subplot_mosaic([[0]], figsize=(4, 4))
Cv = kB * (Theta_vib / T)**2 * np.exp(Theta_vib / T) / (np.exp(Theta_vib / T) - 1)**2
axs[0].plot(Tr, Cv / kB, "k-")
axs[0].annotate(
'$C_V \\rightarrow 0$', xy=(0.15, 0), xytext=(Tr[-1] / 3, 0.1),
arrowprops=dict(arrowstyle='->', color='b'),
bbox=dict(boxstyle='round,pad=0.3', fc='w', ec='b'),
ha='center', va='center', color='b'
)
high_T = axs[0].plot([0, Tr[-1]], [1, 1], "r--", label="$C_V \\rightarrow k_{\\text{B}}$")
labelLines(high_T, zorder=2.5)
axs[0].set_xlabel(r"$T / \Theta_{\text{vib}}$")
axs[0].set_ylabel(r"$C_V / k_{\text{B}}$")
plt.tight_layout()
glue("harmonic_oscillator_heat_capacity", fig, display=False)
plt.close(fig)
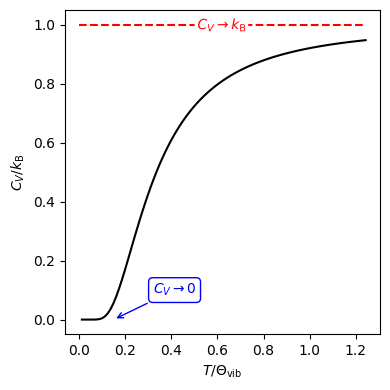
Fig. 19 Heat capacity at constant volume of a harmonic oscillator as a function of temperature. The low-temperature limit is shown as a blue arrow, while the high-temperature limit is shown as a red dashed line.#
Einstein Model#
The Einstein model is a simple model for an atomic crystal, where \(N\) identical atoms are situated at lattice sites and vibrate (each with the same frequency) about these sites as independent (i.e., non-interacting), three-dimensional harmonic oscillators. Since the atoms are situated at lattice sites, they can be treated as distinguishable. For the Einstein model, the partition function is
One can show that
and
Dulong-Petit Law#
Show code cell source
# https://en.wikipedia.org/wiki/Heat_capacities_of_the_elements_(data_page)
import pandas as pd
df = pd.read_csv("../_static/chapter-02/section-06/dulong_petit.csv")
df = df[df.Source == "use"].copy()
df["Atomic Number"] = df["Atomic Number"].astype(int)
df["Molar (J/mol·K)"] = df["Molar (J/mol·K)"].astype(float)
df["Molar (R)"] = df["Molar (J/mol·K)"] / 8.314
df["Absolute Deviation"] = np.abs(df["Molar (R)"] - 3)
# Remove elemental gases
elemental_gases = ["H", "He", "N", "O", "F", "Ne", "Cl", "Ar", "Kr", "Xe", "Rn"]
df = df[~df.Symbol.isin(elemental_gases)]
fig, axs = plt.subplot_mosaic([[0]], figsize=(4, 4))
axs[0].scatter(df["Atomic Number"], df["Molar (R)"], color="black")
axs[0].set_xlabel("Atomic Number")
axs[0].set_ylabel(r"Heat Capacity ($k_{\text{B}}$)")
# Annotate outliers
outliers = df[df["Absolute Deviation"] > 1]
for _, row in outliers.iterrows():
axs[0].annotate(row["Symbol"], (row["Atomic Number"], row["Molar (R)"]))
# Plot Dulong-Petit law
dulong_petit = axs[0].plot([0, 100], [3, 3], "r--", label="Dulong-Petit law")
labelLines(dulong_petit, zorder=2.5)
plt.tight_layout()
glue("dulong_petit", fig, display=False)
plt.close(fig)
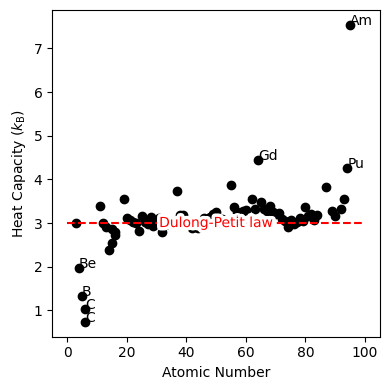
Fig. 20 Heat capacity of elemental solids as a function of atomic number. The Dulong-Petit law is shown as a red dashed line.#
